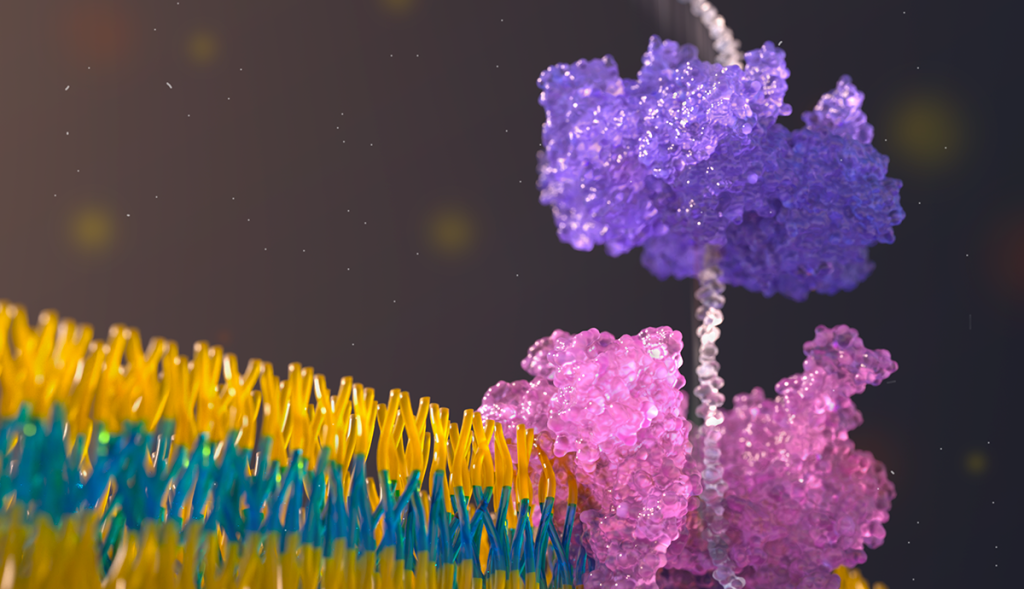
Determining protein sequences, or the order that amino acids are arranged within a protein molecule, is key to understanding their role in different biological processes and diseases. However, current methods for protein sequencing including mass spectrometry are limited and may not be sensitive enough to capture all the varying combinations of molecules in their entirety.
In a recent paper published in the journal Nature, a team of University of Washington researchers introduced a new approach to long-range, single-molecule protein sequencing using commercially available devices from Oxford Nanopore Technologies (ONT). The team, led by senior author and Allen School research professor Jeff Nivala, demonstrated how to read each protein molecule by pulling it through a nanopore sensor. Nanopore technology uses ionic currents that flow through small nanometer-sized pores within a membrane, enabling the detection of molecules that pass through the pore. This can be done multiple times for the same molecule, increasing the sequencing accuracy.
The approach has the potential to help researchers gain a clearer picture of what exists at the protein level within living organisms.
“This research is a foundational advance towards the holy grail of being able to determine the sequence of individual full-length proteins,” said Nivala, co-director of the Molecular Information Systems Lab (MISL).
The technique uses a two-step approach. First, an electrophoretic force pushes the target proteins through a CsgG protein nanopore. Then, a molecular motor called a ClpX unfoldase pulls and controls the translocation of the protein back through the nanopore sensor. Giving each protein multiple passes through the sensor helps eliminate the “noise associated with a single read,” Nivala explained. The team is then able to take the average of all the passes to get a more accurate sequencing readout as well as a detailed detection of any amino acid substitutions and post-translational modifications across the long protein strand.
This method differs from mass spectrometry, which does not look at each individual molecule, but takes the average of an ensemble of different proteins to characterize the sample — potentially losing out on information as each protein can have multiple variations within a cell, Nivala noted.
“One major advantage of nanopore technology is its ability to read individual molecules. However, analyzing these signals at the single-molecule level is challenging because of the variability in the signals, which persist to some extent even after applying normalization and alignment algorithms,” said co-lead author Daphne Kontogiorgos-Heintz, an Allen School Ph.D. student who works with Nivala in the MISL. “This is why I am so excited that we found a method to reread the same molecule multiple times.”
With a more detailed understanding of the protein sequences, this technology can help researchers develop medications that can target specific proteins, tackling cancer and neurological diseases like Alzheimer’s, Nivala explained.
“This will shed light into new diagnostics by having the ability to determine new biomarkers that might be associated with disease that currently we’re not able to to to read,” Nivala said. “It will also develop more opportunities to find new therapeutic targets, because we can find out which proteins could be manifesting the disease and be able to now target those specific variants.”
While the technology can help analyze natural biological proteins, it can also help read synthetic protein molecules. For example, synthetic protein molecules could be designed as data storage devices to record the molecular history of the cell, which would not be possible without the detailed readings from nanopore sensing, Nivala explained. The next step for this research would be working toward increasing the accuracy and resolution to achieve de novo sequencing of single molecule proteins using nanopores, which does not require a reference database.
Nivala and the team were able to conduct this research by modifying ONT technology toward nanopore protein sequencing.
“This study highlights the remarkable versatility of the Oxford Nanopore sensing platform,” said Lakmal Jayasinghe, the company’s SVP of R&D Biologics. “Beyond its established use in sequencing DNA and RNA, the platform can now be adapted for novel applications such as protein sequencing. With its distinctive features including portability, affordability and real-time data analysis, researchers can delve into proteomics at an unprecedented level by performing sequencing of entire proteins using the nanopore platform. Exciting developments lie ahead for the field of proteomics with this groundbreaking advancement.”
Additional authors include former postdoc Keisuke Motone, current Ph.D. student Melissa Queen and current Master’s student Sangbeom Yang (B.S. ‘24) of the Allen School; MISL undergraduate researchers Jasmine Wee, Yishu Fang and Kyoko Kurihara, lab manager Gwendolin Roote and research scientist Oren E. Fox; UW Molecular Engineering and Science Institute Ph.D. student Mattias Tolhurst and Ph.D. alum Nicolas Cardozo; and Miten Jain, a professor of bioengineering and physics at Northeastern University.
Read the full paper in the journal Nature.