Tree-based machine learning models are among the most popular non-linear predictive learning models in use today, with applications in a variety of domains such as medicine, finance, advertising, supply chain management, and more. These models are often described as a “black box” — while their predictions are based on user inputs, how the models arrived at their predictions using those inputs is shrouded in mystery. This is problematic for some use cases, such as medicine, where the patterns and individual variability a model might uncover among various factors can be as important as the prediction itself.
Now, thanks to researchers in the Allen School’s Laboratory of Artificial Intelligence for Medicine and Science (AIMS Lab) and UW Medicine, the path from inputs to predicted outcome has become a lot less dense. In a paper published today in the journal Nature Machine Intelligence, the team presents TreeExplainer, a novel set of tools rooted in game theory that enables exact computation of optimal local explanations for tree-based models.
While there are multiple ways of computing global measures of feature importance that gauge their impact on the model as a whole, TreeExplainer is the first tractable method capable of quantifying an input feature’s local importance to an individual prediction while simultaneously measuring the effect of interactions among multiple features using exact fair allocation rules from game theory. By precisely computing these local explanations across an entire dataset, the tool also yields a deeper understanding of the global behavior of the model. Unlike previous methods for calculating local effects that are impractical or inconsistent when applied to tree-based models and large datasets, TreeExplainer produces rapid local explanations with a high degree of interpretability and strong consistency guarantees.
“For many applications that rely on machine learning predictions to guide decision-making, it is important that models are both accurate and interpretable — meaning we can understand how a model combined and weighted the various input features in predicting a certain result,” explained lead author and recent Allen School alumnus Scott Lundberg (Ph.D., ‘19), now a senior researcher at Microsoft Research. “Precise local explanations of this process can uncover patterns that we otherwise might not see. In medicine, factors such as a person’s age, sex, blood pressure, and body mass index can predict their risk of developing certain conditions or complications. By offering a more robust picture of how these factors contribute, our approach can yield more actionable insights, and hopefully, more positive patient outcomes.”
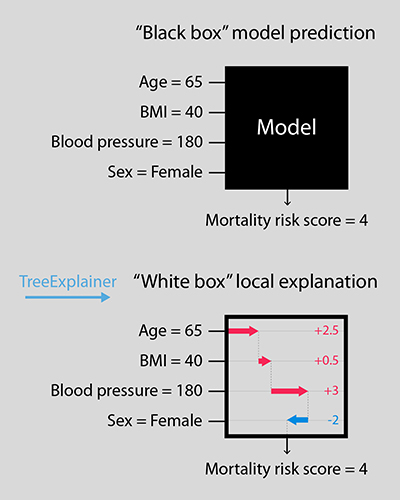
Lundberg and his colleagues offer a new approach to attributing local importance to input features in trees that is both principled and computationally efficient. Their method draws upon game theory to calculate feature importance as classic Shapley values, reducing the complexity of the calculation from exponential to polynomial time to produce explanations that are guaranteed to always be both locally accurate and consistent. To capture interaction effects, the team introduces Shapley Additive Explanation (SHAP) interaction values. These offer a new, richer type of local explanation that employs the Shapley interaction index — a relatively recent concept in game theory — to produce a matrix of feature attributions with uniqueness guarantees similar to Shapley values.
This dual approach enables separate consideration of the main contributions and the interaction effects of features that lead to an individual model prediction, which can uncover patterns in the data that may not be immediately apparent. By combining local explanations from across an entire dataset, TreeExplainer offers a more complete global representation of feature performance that both improves the detection of feature dependencies and succinctly shows the magnitude, prevalence, and direction of each feature’s effect — all while avoiding the inconsistency problems inherent in previous methods.
In a clinical setting, TreeExplainer can provide a global view of the dependency of certain patient risk factors while also highlighting variabilities in individual risk. In their paper, the UW researchers describe several new methods they developed that make use of the local explanations from TreeExplainer to capture global patterns and glean rich insights into a model’s behavior, using multiple medical datasets. For example, the team applied a technique called local model summarization to uncover a set of rare but high-magnitude risk factors for mortality. These are inputs such as high blood protein that are shown to have low global importance, and yet they are extremely important for some individuals’ mortality risk. Another experiment in which the researchers analyzed local interactions for chronic kidney disease revealed a noteworthy connection between high white blood cell counts and high blood urea nitrogen; the team found that the model assigned higher risk to the former when it was accompanied by the latter.
In addition to discerning these patterns, the researchers were able to identify population sub-groups that shared mortality-related risk factors and complementary diagnostic indicators for kidney disease using a technique called local explanation embeddings. In this approach, each sample is embedded into a new “explanation space” to enable supervised clustering in which samples are grouped together based on their explanations. For the mortality dataset, the experiment revealed certain sub-groups within the broader age groups that share specific risk factors, such as younger individuals with inflammation markers or older individuals who are underweight, that would not be apparent using a simple unsupervised clustering method. Unsupervised clustering also would not have revealed how two of the strongest predictors of end-stage renal disease — high blood creatinine levels, and a high ratio of urine protein to urine creatinine — can each be used to identify a set of unique at-risk individuals and should be measured in parallel.

Beyond revealing new patterns of patient risk, the team’s approach also proved useful for exercising quality control over the models themselves. To demonstrate, the researchers monitored a simulated deployment of a hospital procedure duration model. Using TreeExplainer, they were able to identify intentionally introduced errors as well as previously undiscovered problems with input features that degraded the model’s performance over time.
“With TreeExplainer, we aim to break out of the so-called black box and understand how machine learning models arrive at their predictions. This is particularly important in settings such as medicine, where these models can have a profound impact upon people’s lives,” observed Allen School professor Su-In Lee, senior author and director of the AIMS Lab. “We’ve shown how TreeExplainer can enhance our understanding of risk factors for adverse health events.
“Given the popularity of tree-based machine learning models beyond medicine, our work will advance explainable artificial intelligence for a wide range of applications,” she said.
Lee and Lundberg co-authored the paper with joint UW Ph.D./M.D. students Gabriel Erion and Alex DeGrave; Allen School Ph.D. student Hugh Chen; Dr. Jordan Prutkin of the UW Medicine Division of Cardiology; Bala Nair of the UW Medicine Department of Anesthesiology & Pain Medicine; and Ronit Katz, Dr. Jonathan Himmelfarb, and Dr. Nisha Bansal of the Kidney Research Institute.
Learn more about TreeExplainer in the Nature Machine Intelligence paper here and the project webpage here.


