Many people have had the experience of being poked in the back by those annoying plastic tags while trying on clothes in a store. That is just one example of radio frequency identification (RFID) technology, which has become a mainstay not just in retail but also in manufacturing, logistics, transportation, health care, and more. And who wouldn’t recognize the series of black and white lines comprising that old grocery-store standby, the scannable barcode? That invention — which originally dates back to the 1950s — eventually gave rise to the QR code, whose pixel patterns serve as a bridge between physical and digital content in the smartphone era.
Despite their near ubiquity, these object tagging systems have their shortcomings: they may be too large or inflexible for certain applications, they are easily damaged or removed, and they may be impractical to apply in high quantities. But recent advancements in DNA-based data storage and computation offer new possibilities for creating a tagging system that is smaller and lighter than conventional methods.
That’s the point of Porcupine, a new molecular tagging system introduced by University of Washington and Microsoft researchers that can be programmed and read within seconds using a portable nanopore device. In a new paper published in Nature Communications, the team in the Molecular Information Systems Laboratory (MISL) describe how dehydrated strands of synthetic DNA can take the place of bulky plastic or printed barcodes. Building on recent developments in nanopore-based DNA sequencing technologies and raw signal processing tools, the team’s inexpensive and user-friendly design eschews the need for access to specialized labs and equipment.
“Molecular tagging is not a new idea, but existing methods are still complicated and require access to a lab, which rules out many real-world scenarios,” said lead author Kathryn Doroschak, a Ph.D. student in the Allen School. “We designed the first portable, end-to-end molecular tagging system that enables rapid, on-demand encoding and decoding at scale, and which is more accessible than existing molecular tagging methods.”
Instead of radio waves or printed lines, the Porcupine tagging scheme relies on a set of distinct DNA strands called molbits — short for molecular bits — that incorporate highly separable nanopore signals to ease later readout. Each individual molbit comprises one of 96 unique barcode sequences combined with a longer DNA fragment selected from a set of predetermined sequence lengths. Under the Porcupine system, the binary 0s and 1s of a digital tag are signified by the presence or absence of each of the 96 molbits.
“We wanted to prove the concept while achieving a high rate of accuracy, hence the initial 96 barcodes, but we intentionally designed our system to be modular and extensible,” explained MISL co-director Karin Strauss, senior principal research manager at Microsoft Research and affiliate professor in the Allen School. “With these initial barcodes, Porcupine can produce roughly 4.2 billion unique tags using basic laboratory equipment without compromising reliability upon readout.”
Although DNA is notoriously expensive to read and write, Porcupine gets around this by presynthesizing the fragments of DNA. In addition to lowering the cost, this approach has the added advantage of enabling users to arbitrarily mix existing strands to quickly and easily create new tags. The molbits are prepared for readout during initial tag assembly and then dehydrated to extend shelf life of the tags. This approach protects against contamination from other DNA present in the environment while simultaneously reducing readout time later.
Another advantage of the Porcupine system is that molbits are extremely tiny, measuring only a few hundred nanometers in length. In practical terms, this means each molecular tag is small enough to fit over a billion copies within one square millimeter of an object’s surface. This makes them ideal for keeping tabs on small items or flexible surfaces that aren’t suited to conventional tagging methods. Invisible to the naked eye, the nanoscale form factor also adds another layer of security compared to conventional tags.

“Unlike existing inventory control methods, DNA tags can’t be detected by sight or touch. Practically speaking, this means they are difficult to tamper with,” explained co-author Jeff Nivala, a research scientist at the Allen School. “This makes them ideal for tracking high-value items and separating legitimate goods from forgeries. A system like Porcupine could also be used to track important documents. For example, you could envision molecular tagging being used to track voters’ ballots and prevent tampering in future elections.”
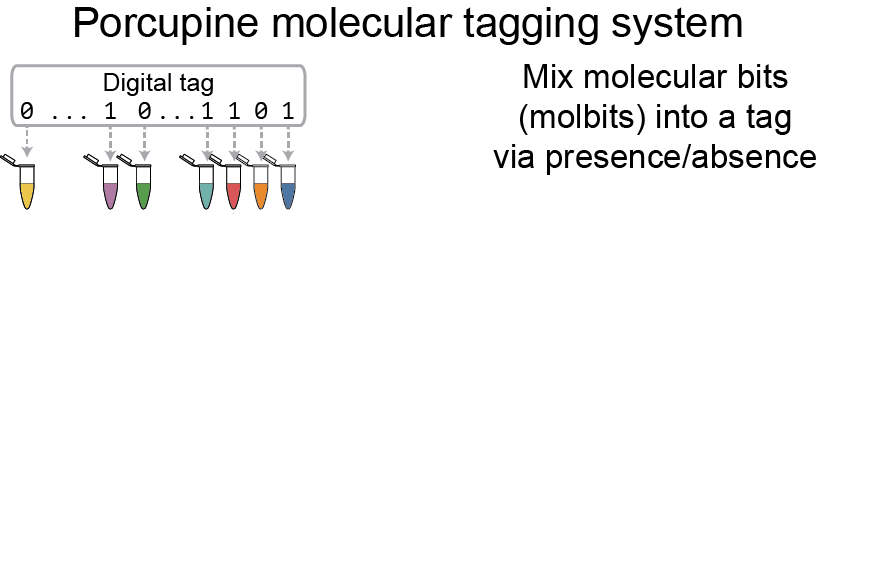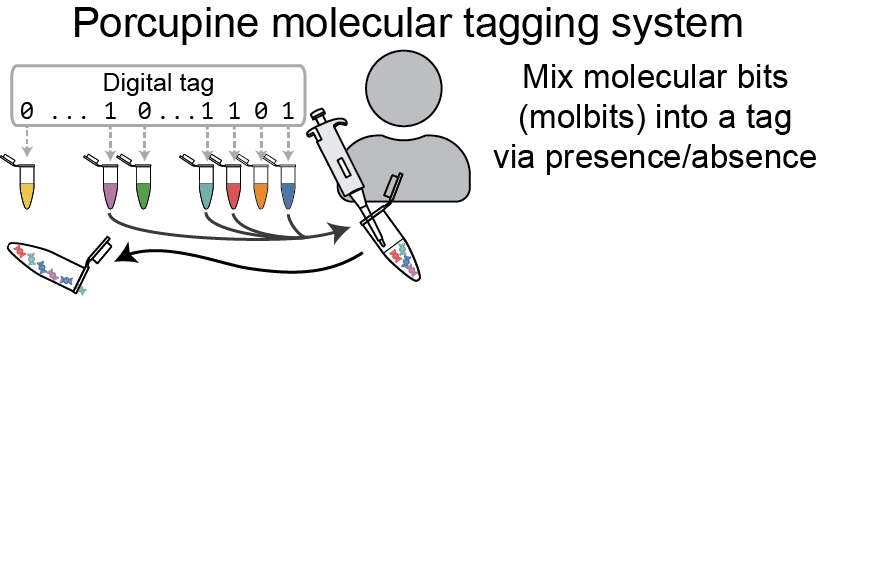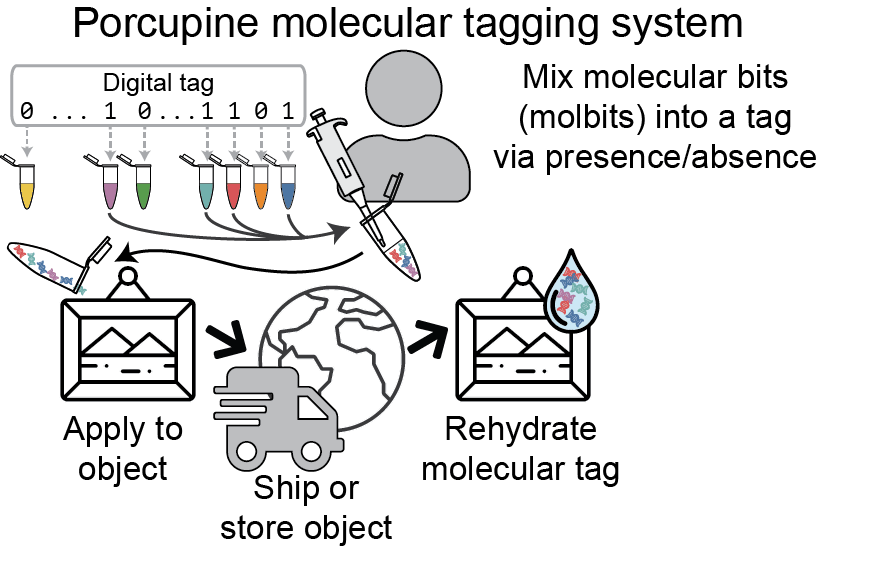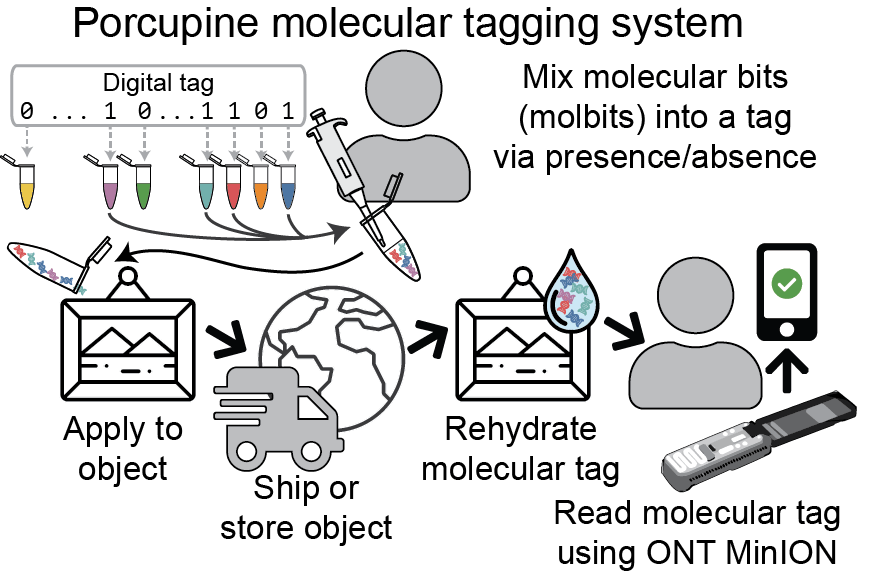
To read the data in a Porcupine tag, a user rehydrates the tag and runs it through a portable Oxford Nanopore Technologies’ MinION device. To demonstrate, the researchers encoded and then decoded their lab acronym, “MISL,” reliably and within a few seconds using the Porcupine system. As advancements in nanopore technologies make them increasingly affordable, the team believes molecular tagging could become an increasingly attractive option in a variety of real-world settings.
“Porcupine is one more exciting example of a hybrid molecular-electronic system, combining molecular engineering, new sensing technology and machine learning to enable new applications.” said Allen School professor and MISL co-director Luis Ceze.
In addition to Ceze, Doroschak, Nivala and Strauss, contributors to the project include Allen School undergraduate Karen Zhang, master’s student Aishwarya Mandyam, and Ph.D. student Melissa Queen. This research was funded in part by the Defense Advanced Research Project Agency (DARPA) under its Molecular Informatics Program and gifts from Microsoft.
Read the paper in Nature Communications here.