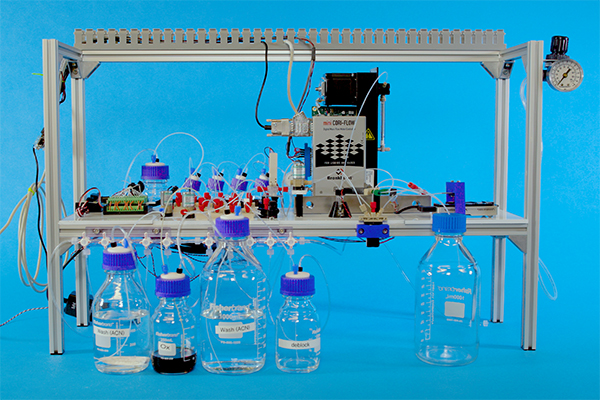
For the past three years, researchers in the Molecular Information Systems Laboratory (MISL) have been on a mission to store the world’s digital data in DNA. A partnership between the University of Washington and Microsoft, the lab has already sparked the imagination of artists, archivists, scientists, and the public with its vision to move beyond traditional data storage media, inspired by the very building blocks of life — what Allen School professor Luis Ceze refers to as “nature’s own perfected storage medium.”
The members of MISL have worked with DNA synthesis company Twist Bioscience to preserve a range of artifacts, from iconic recordings of the Montreux Jazz Festival, to the Universal Declaration of Human Rights, to your neighbor’s favorite cat photo. Along the way, they used these projects to practice encoding and decoding snippets of digital data in synthetic DNA and demonstrate new capabilities for random access and content-based search. They even revealed plans to launch a DNA-based archive into space next year as part of Arch Mission’s Lunar Library, a unique challenge that will compel the researchers to find a way to protect their precious cargo in the harshest of environments for the sake of posterity.
Now the team, co-led by Ceze and Microsoft Principal Researcher Karin Strauss, is ramping up the innovation — and the scientific “wow” factor — with a series of new projects that have opened up exciting new avenues for exploration at the intersection of biology and computer science. One of those projects, described in detail in a new paper published in the journal Nature Communications last week, may be the clearest indicator yet that a DNA-based storage system is not only an intriguing option for solving the world’s data crunch, but also a practical one.
“Once we outlined a DNA storage system, we began contemplating the practical considerations,” said Strauss, who is also an affiliate professor in the Allen School. “The first milestone was figuring out random access within a single pool of DNA molecules mixed together, to retrieve only the data we want and avoid the time and expense of sequencing what we don’t. Our next challenge was to figure out how to take full advantage of DNA’s incredible density and resiliency while automating as many stages of the process as possible. Our latest step shows how to physically organize multiple DNA pools and retrieve them with liquid droplets controlled digitally.”
In their latest paper, Strauss and her colleagues presented a system for achieving high-density data storage in synthetic DNA. By “high-density” they mean one full terabyte of data — the equivalent of 1,000 gigabytes — in a single spot of dehydrated DNA one millimeter in diameter, or roughly the size of a pinhead. Although the information density of DNA molecules is theoretically much higher than that, the team wanted to ensure the ability to retrieve specific data from a particular pool without having to sequence the entire pool.
The team arranged the spots of DNA on glass cartridges, with each cartridge capable of storing up to 50 terabytes based on current DNA storage techniques. Multiple glass cartridges can then be stacked in a space-saving vertical configuration — akin to the approach taken with existing magnetic tape or hard drive-based storage systems to conserve room, albeit much more compact.
“DNA must be in liquid form for sample preparation and sequencing, but isolating liquid samples can be a cumbersome process and requires separate vessels — which would sacrifice a significant amount of density,” explained lead author Sharon Newman, an alumna of UW Bioengineering who is currently pursuing a Ph.D. at Stanford University. “Our dry storage architecture enables us to store data with much higher density compared to other approaches, and allows for physical isolation and data retrieval without risking contamination from other samples.”
To retrieve the data, the researchers rehydrate the DNA with a droplet of water using a digital microfluidics (DMF) device. Newman and her colleagues took a keen interest in DMF technology, which is capable of manipulating liquids in very small quantities with higher precision than humans. By automating biological and chemical protocols with DMF, researchers can scale up the processes involved in implementing DNA data storage. The devices are particularly suited to DNA storage processing, but they have their limitations: not only do DMF platforms tend to be prohibitively expensive, but they are also inflexible, error-prone, and difficult to program.
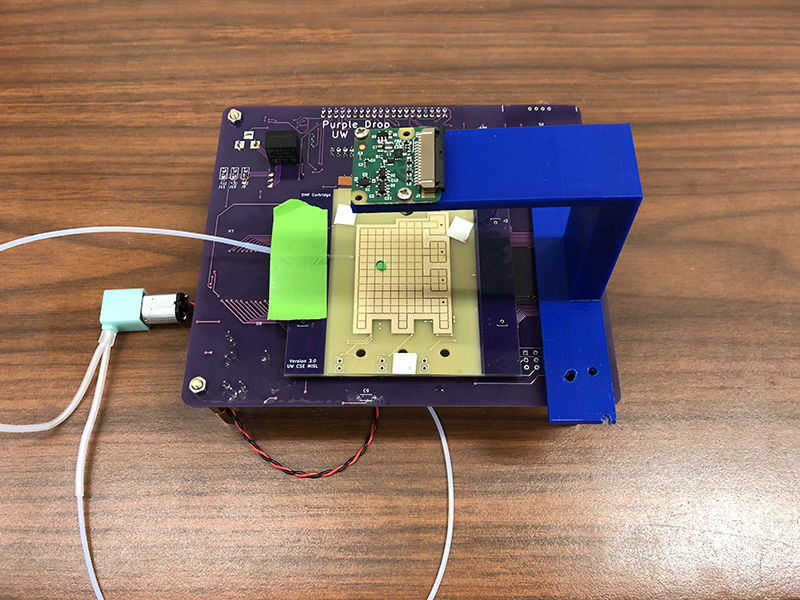
Recognizing that DNA data storage would remain a pipe dream as long as the combination of cost and complexity made it inaccessible to all but a handful of experts in resource-rich laboratories, a group of MISL researchers developed a low-cost, general-purpose DMF device for holding and manipulating the droplets of DNA, which they dubbed PurpleDrop, and a full-stack DMF automation platform known as Puddle. Together, the projects — which the team will present at the Association for Computing Machinery’s 24th International Conference on Architectural Support for Programming Languages and Operating Systems (ASPLOS 2019) this week — comprise a complete DMF programming system that will make microfluidic technology more accessible while simultaneously expanding its computational capabilities.
“Most of the work on microfluidics has focused on automating individual protocols, in which the device is given a fixed set of inputs, and manipulates them in a specified way to produce an output,” explained Allen School Ph.D. student Max Willsey. “Although this is an important component of wet-lab research, it limits the role of DMF to that of a microcontroller. We envision a more expansive and dynamic system that enables scientists to program complex protocols in Python or another language of choice while providing real-time error correction.”
Puddle’s dynamic approach to resource management sets it apart from existing techniques, which take a more static approach to microfluidic programming. Puddle is an application programming interface (API) that purposefully maximizes expressiveness and ease of use, in exchange for sacrificing some efficiency and ahead-of-time guarantees. This trade-off gives Puddle more flexibility, allowing both the system and the user to react to data from the fluidic domain. These data-driven decisions fall into three categories: protocol-level decisions, such as automatic replenishment of a liquid that has evaporated during an experiment; application-level decisions based on the protocol output, such as what experiment to run next; and execution-level decisions, such as error detection and correction. To enable the latter, the team employed computer vision and a small camera mounted on top of the DMF device. The camera functions as a multi-purpose sensor for detecting the location and volume of droplets to help Puddle decide if an error has occurred.

The built-in error detection enables the robust execution of the system on relatively cheap hardware, meaning the researchers could prioritize simplicity and accessibility in designing the PurpleDrop device. In addition to the camera, PurpleDrop features a Raspberry Pi 3B single-board computer, instead of a microcontroller, to drive the electronics, which enables it to function as a self-sufficient microfluidics platform. Costing about$300 assembled — orders of magnitude less than most fluidics systems — the design is also simple enough for many labs to put together on their own, without requiring access to a clean room.
“Cost considerations are one of the main sticking points when it comes to microfluidics,” noted Allen School research scientist and co-author Ashley Stephenson. “So as we look for ways to expand the capabilities, we want to ensure that scientists and practitioners will be able to access these innovations. This work can be used to advance not just DNA data storage but also many other areas of research, such as medicine.”
Because cost and complexity are probably the two biggest barriers to widespread adoption of DNA as a storage medium, it comes as no surprise that automation has emerged as a recurring theme in MISL’s work. Last month, the world said “hello” to the first fully automated, end-to-end system for storing digital data in synthetic DNA. Lab members took those five letters, represented by five bytes of data, and ran them through a fully functioning prototype incorporating the equipment required to encode, synthesize, pool, sequence, and read back the data — the majority of which, like PurpleDrop, was built using inexpensive, off-the-shelf components. And it performed the cycle without human intervention, which as senior research scientist Chris Takahashi pointed out, will be an advantage when it comes to DNA data storage in the wild.
“You can’t have a bunch of people running around a data center with pipettes,” pointed out Takahashi, lead author of a related paper published in Nature Scientific Reports. “It’s too prone to human error, too costly, and the footprint would be too large.”
Takahashi and his colleagues did not set out to demonstrate speed or even affordability at this stage; rather, they built the machine to show that end-to-end automation was possible. The team’s ultimate goal is to develop a system that resembles any other cloud-based storage service, to which end users would be able to upload their data to a storage center. The difference is, instead of staying in digital form, a customer’s data would be converted to the As, Ts, Cs, and Gs of DNA until it is needed again.
“We are developing an entirely new way of storing digital data from scratch, which means building all new hardware, platforms, and techniques,” said Ceze. “There is a lot more to it than solving the technical challenges related to converting those 0s and 1s to DNA molecules, and we have made significant progress in the last three years. With these latest results, we are building a bridge between computation and molecular biology and introducing exciting new capabilities that will benefit both fields.”
The Nature Communications paper on high-density DNA data storage was co-authored by Newman, Stephenson, Willsey, Takahashi, Strauss, Ceze, and Microsoft researcher Bichlien Nguyen.
The ASPLOS paper on Puddle and PurpleDrop was co-authored by Willsey, Stephenson, Takahashi, Nguyen, Newman, Strauss, Ceze, high school intern Pranav Vaid, Allen School master’s students Michal Piszczek and Christine Betts, and Allen School alumnus Sarang Joshi (B.S., ‘18).
The Nature Scientific Reports paper on end-to-end automation of DNA data storage was co-authored by Takahashi, Nguyen, Strauss, and Ceze.
To learn more about the DNA data storage research, visit the MISL website here.